הפקולטה לביולוגיה בטכניון היא קהילה דינמית של סטודנטים וחוקרים, הנהנת ממעטפת של מוסד טכנולוגי מוביל. אנו מציעים חוויית למידה מיוחדת, המשלבת תוכניות אקדמיות מעמיקות עם התנסות במחקר מעשי, יחס סגל-סטודנטים גבוהה המבטיח תשומת לב אישית ומקדם את החשיבה הביקורתית למנהיגות מדעית והתמודדות עם האתגרים המדעיים של המאה ה-21.
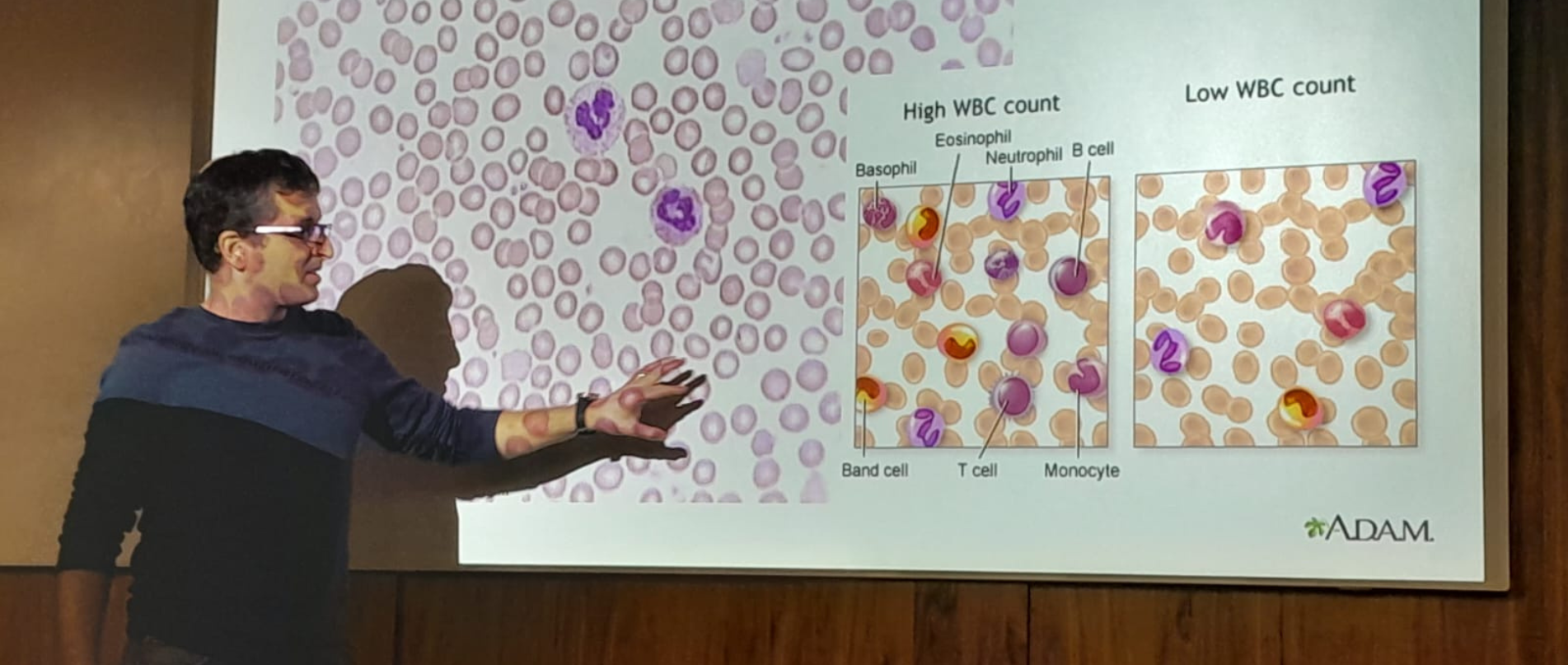
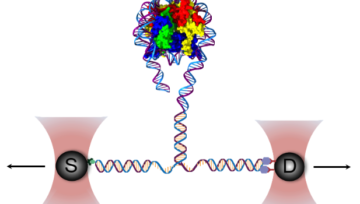
צוותי המחקר שלנו שואלים שאלות מקוריות ומורכבות המכסות מגוון רחב של תחומי דעת, לדוגמא: עמידות אנטיביוטית בחיידקים, התמצאות החיה או הצמח במרחב, מקור מחלת האלצהיימר, פיתוח תרופות לסרטן, שימוש באנרגיה פוטו-אלקטרית על ידי ווירוסים וחיידקים, סימביוזה ואבולוציה. כיאה למכון טכנולוגי, הפתרונות מתקבלים באמצעות מערכות פיזיקליות מתקדמות במערכות ניסוייות מגוונות.
בכדי לטפח רעיונות מקוריים, אנו פותחים את שערינו בפני סטודנטים וחוקרים מרחבי העולם מרקעים ותרבויות שונות ומספקים סביבה המטפחת סקרנות, שיתוף פעולה ותמיכה הדדית. הסגל והסטודנטים שלנו הינם מדענים בעלי שם והבוגרים שלנו מתברגים בתפקידי מפתח באקדמיה ותעשיית הביוטכנולוגיה בארץ ובעולם.
בשנים הקרובות, אנחנו מתכננים להתרחב לחזיתות מחקר חדשות, לחזק שותפויות גלובליות עם האקדמיה והתעשייה ולהמשיך לדחוף את גבולות הידע. בין אם א או אתה מתעניינים בתואר אקדמי, או כבר חוקרים מנוסים, אנו מזמינים אתכם להצטרף אלינו למסע מרתק לגלות את רזי החיים ולייצר אימפקט משמעותי על החברה.
דיקן הפקולטה פרופ' מיכאל גליקמן