SPOTLIGHT ON RESEARCH
PAPER OF THE MONTH SEP 21
The Committee for Graduate Studies in the Faculty of Biology selects each month the leading scientific article from all the scientific articles published for that month.
We are pleased to announce that the winner of this September’s article is the doctoral student Dror Shitrit from Prof. Debbie Lindell’s laboratory, whose article was published in The ISME Journal.
On the occasion of the win, we asked Dror to provide us with some interesting details about the study, the path that led to the research and also a bit about himself.
Hi Dror, could you introduce yourself in a few words?
I am a microbiologist, currently a post-doc at the Lindell lab. I live in Haifa with my wife, Gal, and we are expecting our first daughter to arrive very soon.
Could you explain what Prof. Lindell’s laboratory is about?
We study marine cyanobacteria, which are bacteria that perform photosynthesis, just like plants. In fact, they are much more ancient than plants and are still highly abundant today in the oceans and other environments, producing a significant portion of the oxygen we breathe. Beyond the cyanobacteria themselves, we are interested in viruses that infect them. These are called cyanophages and are also highly abundant where cyanobacteria are found. By infecting and killing cyanobacteria, they have a major influence on cyanobacterial populations, as well as on the global biogeochemical cycle of matter. We study these bacteria and viruses using lab experiments with strains that were isolated from the sea and we grow in the lab, and also by performing field experiments. We participate in scientific cruises quite often, both in the Israeli Red Sea and in the oceans across the world.
Could you tell us about your current article/research, what was the main purpose of the research and what did you discover?
Our paper first introduces new genetic engineering that enables modification of the genomes of cyanophages and potentially other viruses. The reason we need such a method is that it is essential in order to investigate the gene content of these viruses and to understand how they work. In the last decade, with the development of DNA sequencing techniques, a lot of genomic data has accumulated, and we know a lot about what genes cyanophage have. What was missing is a way to test what these genes do. In some cases, we have no clue because no similar genes are found in the existing databases. In other cases, it is the resemblance to known genes that is puzzling. Some cyanophage genes have predicted functions that don’t make a lot of sense for a virus to have, like photosynthesis genes for example. Once we developed the method we used it to address such a question, regarding the presence of lysogeny-associated genes in T7-like cyanophages. Lysogeny is a phenomenon in which a virus remains in a dormant state inside the host it infects, typically after integrating its whole genome into the host’s genome. Some cyanophages that we have in the lab have genes that are predicted to perform this genome integration, but when we grow them in the lab they don’t appear as lysogenic phages. What we found is that although they are able to insert their genome into the host genome, they only do it transiently and this is not a part of classic lysogeny.

Various species of cyanobacteria isolated from the ocean
Can you elaborate on the importance of the discovery? Why will this discovery serve you and what directions does it take? What is the application of the discovery (domains, solutions)? The method enables the direct characterization of cyanophage genes for the first time. Until now they relied largely on taking viral DNA sequences from field samples and lab isolated and comparing them to the existing database. This approach is useful for getting an initial understanding of an organism, but it doesn’t leave much room for new discoveries. It’s important to note that studying other viruses that infect bacteria has led to countless important discoveries, from basic mechanisms in cell biology to cutting-edge genetic engineering methods such as CRISPR-Cas methods. What we found in our paper is, in my view, a good example of the limitations of making conclusions based on sequence content alone. We hope that our results and method will start a shift toward experiment-based conclusions (where it is possible, of course), and will lead to additional new discoveries.
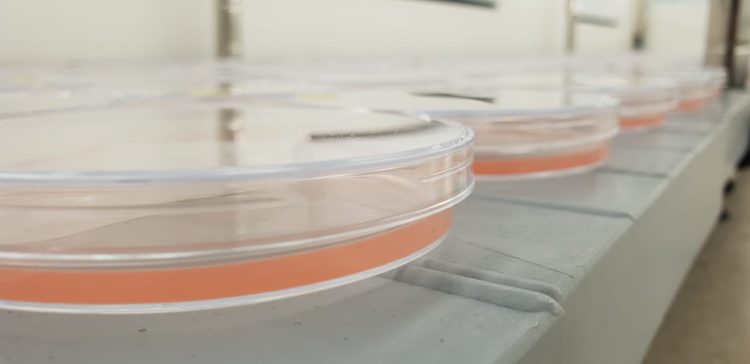
Cyanobacteria (Synechococcus) grow in Petri dishes
Can you tell us a little about the work process / how long did it take to work on the current study? Do you start an article when you know where you want to go? What happens next (corona, unplanned disturbances, how does it affect things?
Developing a new method or an experimental setup from scratch is usually a very long process, and this was the case here too. My first idea was to take a method that I developed in my previous lab (a CRISPR-Cas-based method) and to modify it for cyanophages. After it didn’t work I tried few other approaches that didn’t work as well. I was very close to giving up but then I decided to use something very simple, almost too simple to work, but it did work on every cyanophage that I tried it on. This process took over two years (cyanobacteria grow very slow, closer to plants than to other lab-grown bacteria), and then I had to decide what gene I want to investigate first. I chose one that seemed like a safe bet, but it wasn’t as easy as I thought to get a good story out of it, and again I had to find original ways to get interesting data. All together this paper is a result of roughly 4 years of work, but not all of this time was spent on this project alone.
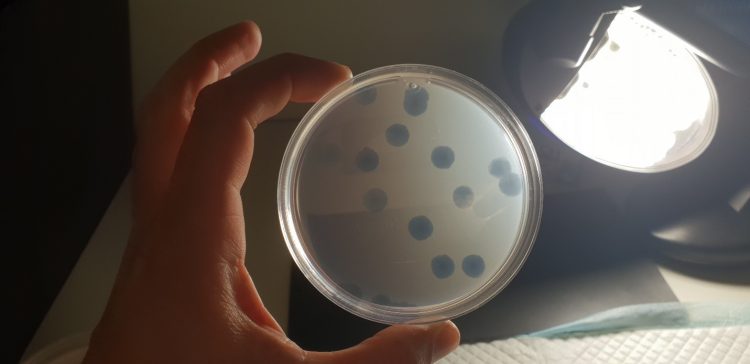
Cyanophages when grown in a laboratory: The plate is full of cyanobacteria (the background).
The round holes called "plaques" are places where Cyanophages have infected and killed the cyanobacteria, so they are transparent.
What drew you to the current lab/project?
I have been interested in cyanobacteria since I learned about them in my B.Sc. courses about the origin of life and general microbiology. I came across phages (viruses that infect bacteria) just by chance due to an unexpected result in my
master’s degree. As I got to know about them I found them to be fascinating organisms, that really take the principles of evolution to the edge, and also stretch our definitions of a living organism. I later worked on phages that infect E. coli bacteria, and although it was interesting, I wanted to study organisms that have a real impact on natural environments, not just on humans. I saw a lecture by Debbie’s student at a conference and did the transition as soon as I had the chance.
A funny/interesting story from the course of the research?
I made a bet with our lab manager, Gazalah, that I will finish my Ph.D. in four years. I still owe her a carrot cake.
When you are not “doing” science, what do you do?
I like hiking, playing football and matkot (not simultaneously…), building, and fixing things. I also really like gardening and I am nurturing our building’s yard for a few years now, growing herbs and vegetables there.
Tell us something no one knows about you…
I wrote the Hebrew Wikipedia entry for “American cockroach”.
When you grow up what do you want to be?
I am quite a grown-up and still not sure about it.
what are your plans for the future of your career?
I would like to keep doing research, in the academy or industry, preferably something with environmental implications.
A tip for a Biology student who started this year?
Go read the textbooks, they are often better than the lectures. In particular, learning about how things were discovered is very interesting and many times helpful.
A link to the full article:https://www.nature.com/articles/s41396-021-01085-8_
A link to Lindell lab site: https://lindelltechnion.wixsite.com/lindell-lab
To Debbie Lindell’s page: https://biology.technion.ac.il/member/%d7%9c%d7%99%d7%a0%d7%93%d7%9c/








