The Committee for Graduate Studies in the Faculty of Biology selects the leading scientific article each month from all the scientific articles published for that month.
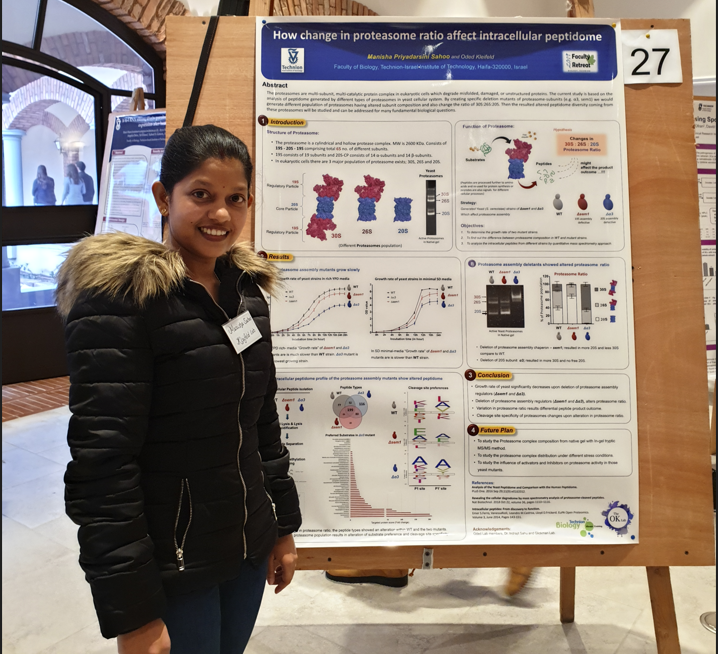
We are pleased to announce that the winner of January article is Manisha Priyadarsini Sahoo from Prof. Kleifeld Laboratory. Alongside Manisha, Dr. Tali Lavy co-authored this manuscript. The article was published in “Molecular and Cellular Proteomics”.
On the occasion of the win, we asked Manisha to provide us with some interesting details about the study, the path that led to the research, and a bit about her.
•Hi, could you introduce yourself in a few words?
Shalom, Basically I am from India and did my second master’s in biology from Oded kleifeld lab, that was the best experience and good exposure I had in a foreign country.
•Could you explain what Prof. Kleifeld Laboratory is about?
OK Lab is the proteomics lab developing new techniques, strategies, and biochemical approaches using mass-spectrometry as the major tool to answer fundamental biological questions.
•Could you tell us about your current article/research what was the main purpose of the research and what did you discover?
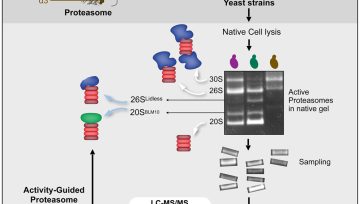
The article’s main purpose is to build a methodology to study the active and inactive proteasome complexes from live eukaryotic cells. Surprisingly we discovered a population of proteasome complex that are matured but lack activity due to the incomplete processing of certain catalytic subunits.
•Can you elaborate on the importance of the discovery? How will it serve you and what directions does it take? What is the application of the discovery (domains, solutions)?
To some extent, we successfully developed an easy, affordable, and quick approach to quantitatively analyze the active pool of proteasome complex isolated from cells. Apart from the methodology, certain interesting findings from our study opens several interesting questions and directions which not only excite me but will also other researchers in the field.
One of the important applications of the study is that our simple proteomic-based methodology can be valuable to investigate other active enzymes and complexes of live cells.
•Name 3 prominent tools that you received in the laboratory during your work and studies?
Learned Native gel technique for large protein complex, Mass spectrometry-based proteomics, and important software and tools (Maxquant, MsFragger, Perseus) to analyze proteomics data.
•When you are not “doing” science, what do you do?
I love to cook and travel.
•What are your plans for the future of your career?
I want to continue my career in proteomic-based research. Wherever I work in the future I will try to implement the knowledge I learned from OK lab.
➡ A link to the full article: https://doi.org/10.1016/j.mcpro.2024.100728
➡ A link to the prof. Kleifeld lab site: https://oklab.net.technion.ac.il/
➡ To Prof. Kleifeld page: https://biology.technion.ac.il/en/member/kleifeld/